.
Adjacency matrix
In mathematics and computer science, an adjacency matrix is a means of representing which vertices (or nodes) of a graph are adjacent to which other vertices. Another matrix representation for a graph is the incidence matrix.
Specifically, the adjacency matrix of a finite graph G on n vertices is the n × n matrix where the non-diagonal entry aij is the number of edges from vertex i to vertex j, and the diagonal entry aii, depending on the convention, is either once or twice the number of edges (loops) from vertex i to itself. Undirected graphs often use the latter convention of counting loops twice, whereas directed graphs typically use the former convention. There exists a unique adjacency matrix for each isomorphism class of graphs (up to permuting rows and columns), and it is not the adjacency matrix of any other isomorphism class of graphs. In the special case of a finite simple graph, the adjacency matrix is a (0,1)-matrix with zeros on its diagonal. If the graph is undirected, the adjacency matrix is symmetric.
The relationship between a graph and the eigenvalues and eigenvectors of its adjacency matrix is studied in spectral graph theory.
Examples
The convention followed here is that an adjacent edge counts 1 in the matrix for an undirected graph.
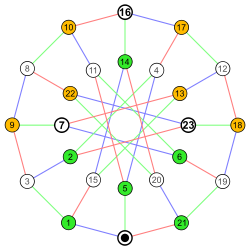
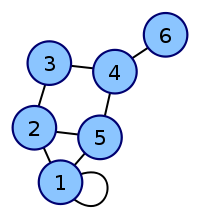
Labeled graph Adjacency matrix
6n-graph2.svg \begin{pmatrix} 1 & 1 & 0 & 0 & 1 & 0\\ 1 & 0 & 1 & 0 & 1 & 0\\ 0 & 1 & 0 & 1 & 0 & 0\\ 0 & 0 & 1 & 0 & 1 & 1\\ 1 & 1 & 0 & 1 & 0 & 0\\ 0 & 0 & 0 & 1 & 0 & 0\\ \end{pmatrix}
| Labeled graph | Adjacency matrix |
|---|---|
 |
\( \begin{pmatrix} 1 & 1 & 0 & 0 & 1 & 0\\ 1 & 0 & 1 & 0 & 1 & 0\\ 0 & 1 & 0 & 1 & 0 & 0\\ 0 & 0 & 1 & 0 & 1 & 1\\ 1 & 1 & 0 & 1 & 0 & 0\\ 0 & 0 & 0 & 1 & 0 & 0\\ \end{pmatrix} \) Coordinates are 1-6. |
|
|
Coordinates are 0-23. |
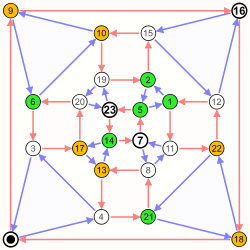
|
As the graph is directed, |
The adjacency matrix of a complete graph is all 1's except for 0's on the diagonal.
The adjacency matrix of an empty graph is a zero matrix.
Adjacency matrix of a bipartite graph
The adjacency matrix A of a bipartite graph whose parts have r and s vertices has the form
\( A = \begin{pmatrix} O & B \\ B^T & O \end{pmatrix}, \)
where B is an r × s matrix and O is an all-zero matrix. Clearly, the matrix B uniquely represents the bipartite graphs. It is sometimes called the biadjacency matrix. Formally, let G = (U, V, E) be a bipartite graph with parts \( U={u_1,..., u_r} and \( V={v_1,..., v_s} \). The biadjacency matrix is the r x s 0-1 matrix B in which \( b_{i,j} = 1 iff (u_i, v_j) \in E \).
If G is a bipartite multigraph or weighted graph then the elements \( b_{i,j} \) are taken to be the number of edges between the vertices or the weight of the edge \( (u_i, v_j) \), respectively.
Properties
The adjacency matrix of an undirected simple graph is symmetric, and therefore has a complete set of real eigenvalues and an orthogonal eigenvector basis. The set of eigenvalues of a graph is the spectrum of the graph.
Suppose two directed or undirected graphs G_1 and G_2 with adjacency matrices \( A_1 \) and \( A_2 \) are given. G_1 and G_2 are isomorphic if and only if there exists a permutation matrix P such that
\( P A_1 P^{-1} = A_2. \)
In particular, A_1 and A_2 are similar and therefore have the same minimal polynomial, characteristic polynomial, eigenvalues, determinant and trace. These can therefore serve as isomorphism invariants of graphs. However, two graphs may possess the same set of eigenvalues but not be isomorphic.
If A is the adjacency matrix of the directed or undirected graph G, then the matrix An (i.e., the matrix product of n copies of A) has an interesting interpretation: the entry in row i and column j gives the number of (directed or undirected) walks of length n from vertex i to vertex j. This implies, for example, that the number of triangles in an undirected graph G is exactly the trace of A3 divided by 6.
The main diagonal of every adjacency matrix corresponding to a graph without loops has all zero entries. Note that here 'loops' means, for example A->A, not 'cycles' such as A->B->A.
For\( \left( d \right) \) -regular graphs, d is also an eigenvalue of A for the vector \( v=\left( 1,\dots,1 \right) \), and G is connected if and only if the multiplicity of d is 1. It can be shown that -d is also an eigenvalue of A if G is a connected bipartite graph. The above are results of Perron–Frobenius theorem.
Variations
An (a, b, c)-adjacency matrix A of a simple graph has Aij = a if ij is an edge, b if it is not, and c on the diagonal. The Seidel adjacency matrix is a (−1,1,0)-adjacency matrix. This matrix is used in studying strongly regular graphs and two-graphs.[1]
The distance matrix has in position (i,j) the distance between vertices vi and vj . The distance is the length of a shortest path connecting the vertices. Unless lengths of edges are explicitly provided, the length of a path is the number of edges in it. The distance matrix resembles a high power of the adjacency matrix, but instead of telling only whether or not two vertices are connected (i.e., the connection matrix, which contains boolean values), it gives the exact distance between them.
Data structures
For use as a data structure, the main alternative to the adjacency matrix is the adjacency list. Because each entry in the adjacency matrix requires only one bit, it can be represented in a very compact way, occupying only \( {n^2} / 8 \) bytes of contiguous space, where n is the number of vertices. Besides avoiding wasted space, this compactness encourages locality of reference.
However, if the graph is sparse, adjacency lists require less storage space, because they do not waste any space to represent edges that are not present. Using a naïve array implementation on a 32-bit computer, an adjacency list for an undirected graph requires about 8 e bytes of storage, where e is the number of edges.
Noting that a simple graph can have at most \( n^2 \) edges, allowing loops, we can let \( d = e / n^2 \) denote the density of the graph. Then,\( 8 e > n^2 / 8 \), or the adjacency list representation occupies more space precisely when d > 1/64. Thus a graph must be sparse indeed to justify an adjacency list representation.
Besides the space tradeoff, the different data structures also facilitate different operations. Finding all vertices adjacent to a given vertex in an adjacency list is as simple as reading the list. With an adjacency matrix, an entire row must instead be scanned, which takes O(n) time. Whether there is an edge between two given vertices can be determined at once with an adjacency matrix, while requiring time proportional to the minimum degree of the two vertices with the adjacency list.
References
^ Seidel, J. J. (1968). "Strongly Regular Graphs with (−1,1,0) Adjacency Matrix Having Eigenvalue 3". Lin. Alg. Appl. 1 (2): 281–298. doi:10.1016/0024-3795(68)90008-6.
Further reading
Cormen, Thomas H.; Leiserson, Charles E.; Rivest, Ronald L.; Stein, Clifford (2001). "Section 22.1: Representations of graphs". Introduction to Algorithms (Second ed.). MIT Press and McGraw-Hill. pp. 527–531. ISBN 0-262-03293-7.
Godsil, Chris; Royle, Gordon (2001). Algebraic Graph Theory. New York: Springer. ISBN 0-387-95241-1.
External links
Fluffschack — an educational Java web start game demonstrating the relationship between adjacency matrices and graphs.
Open Data Structures - Section 12.1 - AdjacencyMatrix: Representing a Graph by a Matrix
McKay, Brendan. "Description of graph6 and sparse6 encodings".
Café math : Adjacency Matrices of Graphs : Application of the adjacency matrices to the computation generating series of walks.
Retrieved from "http://en.wikipedia.org/"
All text is available under the terms of the GNU Free Documentation License