A primer is a strand of nucleic acid that serves as a starting point for DNA synthesis. They are required because the enzymes that catalyze replication, DNA polymerases, can only add new nucleotides to an existing strand of DNA. The polymerase starts replication at the 3'-end of the primer, and copies the opposite strand. In most cases of natural DNA replication, the primer for DNA synthesis and replication is a short strand of RNA (which can be made de novo). Many of the laboratory techniques of biochemistry and molecular biology that involve DNA polymerase, such as DNA sequencing and the polymerase chain reaction (PCR), require DNA primers. These primers are usually short, chemically synthesized oligonucleotides, with a length of about twenty bases. They are hybridized to a target DNA, which is then copied by the polymerase. Mechanism in vivo
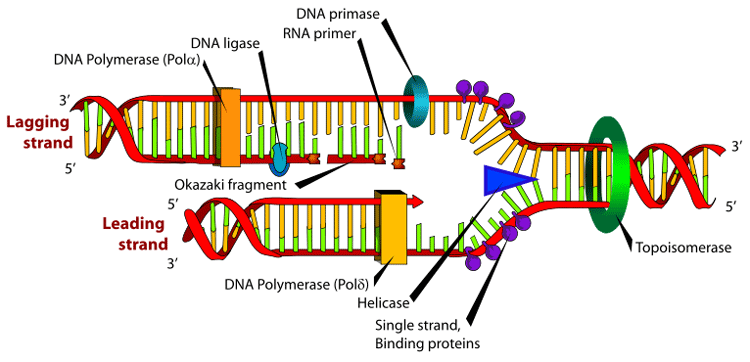
The DNA replication fork. RNA primer labeled at top. Further information: DNA replication, The replication fork The lagging strand is that strand of the DNA double helix that is orientated in a 5' to 3' manner. Because DNA polymerase III cannot synthesize in the 3'→5' direction, the lagging strand is synthesized in short segments known as Okazaki fragments. Along the lagging strand's template, primase builds RNA primers in short bursts. DNA polymerases are then able to use the free 3'-OH groups on the RNA primers to synthesize DNA in the 5'→3' direction. The RNA fragments are then removed by DNA polymerase I for prokaryotes or DNA polymerase δ for eukaryotes (different mechanisms are used in eukaryotes and prokaryotes) and new deoxyribonucleotides are added to fill the gaps where the RNA was present. DNA ligase then joins the deoxyribonucleotides together, completing the synthesis of the lagging strand. Primer removal In eukaryotic primer removal, DNA polymerase δ extends the Okazaki fragment in 5' to 3' direction, and when it encounters the RNA primer from the previous Okazaki fragment, displacing the 5′ end of the primer into a single-stranded RNA flap, which is removed by nuclease cleavage. Cleavage of the RNA flaps involves either endonuclease 1 (FEN1) cleavage of short flaps, or coating of long flaps by the single-stranded DNA binding protein replication protein A (RPA) and sequential cleavage by Dna2 nuclease and FEN1.[1] This mechanism is a potential explanation to how HIV virus can transform its genome into double stranded DNA from the RNA-DNA formed after reverse transcription of its RNA. However, the HIV-encoded reverse transcriptase has own ribonuclease activity that degrades the viral RNA during the synthesis of cDNA, as well as DNA-dependent DNA polymerase activity that copies the sense cDNA strand into a antisense DNA to form a double-stranded DNA intermediate.[2] Uses of synthetic primers DNA sequencing is used to determine the nucleotides in a DNA strand; the chain termination method (dideoxy sequencing or Sanger method) uses a primer as a start marker for the chain reaction. In PCR, primers are used to determine the DNA fragment to be amplified by the PCR process. The length of primers is usually not more than 30 (usually 18-24[3]) nucleotides, and they need to match the beginning and the end of the DNA fragment to be amplified. They direct replication towards each other - the extension of one primer by polymerase then becomes the template for the other, leading to an exponential increase in the target segment. It is worth noting that primers are not essentially always necessary for DNA synthesis and can in fact be used by viral polymerases, e.g. influenza, for RNA synthesis. PCR primer design Pairs of primers should have similar melting temperatures since annealing in a PCR occurs for both simultaneously. A primer with a Tm significantly higher than the reaction's annealing temperature may mishybridize and extend at an incorrect location along the DNA sequence, while Tm significantly lower than the annealing temperature may fail to anneal and extend at all. A good Tm calculator can be found in Finnzymes Reagents site: https://www.finnzymes.fi/tm_determination.html Primer sequences need to be chosen to uniquely select for a region of DNA, avoiding the possibility of mishybridization to a similar sequence nearby. A commonly used method is BLAST search whereby all the possible regions to which a primer may bind can be seen. Both the nucleotide sequence as well as the primer itself can be BLAST searched. The free NCBI tool Primer-BLAST integrates primer design tool and BLAST search into one application. Alternatively use of software such as Beacon designer, may yield to specific primers. Mononucleotide repeats should be avoided, as loop formation can occur and contribute to mishybridization. Primers should not easily anneal with other primers in the mixture (either other copies of same or the reverse direction primer); this phenomenon can lead to the production of 'primer dimer' products contaminating the mixture. Primers should also not anneal strongly to themselves, as internal hairpins and loops could hinder the annealing with the template DNA. When designing a primer for use in TA cloning, efficiency can be increased by adding AG tails to the 5' and the 3' end.[4] Remember that the reverse primer has to be the reverse complement of the given cDNA sequence. The reverse complement can be easily determined in http://www.bioinformatics.org/sms/rev_comp.html Degenerate primers Sometimes degenerate primers are used. These are actually mixtures of similar, but not identical primers. They may be convenient if the same gene is to be amplified from different organisms, as the genes themselves are probably similar but not identical. The other use for degenerate primers is when primer design is based on protein sequence. As several different codons can code for one amino acid, it is often difficult to deduce which codon is used in a particular case. Therefore primer sequence corresponding to the amino acid isoleucine might be "ATH", where A stands for adenine, T for thymine, and H for adenine, thymine, or cytosine, according to the genetic code for each codon, using the IUPAC symbols for degenerate bases. Use of degenerate primers can greatly reduce the specificity of the PCR amplification. The problem can be partly solved by using touchdown PCR. Degenerate primers are widely used and extremely useful in the field of microbial ecology. They allow for the amplification of genes from thus far uncultivated microorganisms or allow the recovery of genes from organisms where genomic information is not available. Usually, degenerate primers are designed by aligning gene sequencing found in GenBank. Differences among sequences are accounted for by using IUPAC degeneracies for individual bases. PCR primers are then synthesized as a mixture of primers corresponding to all permutations. See also Oligonucleotide synthesis, the methods by which primers are manufactured External links * manual of primer generation
1. ^ Distinguishing the pathways of primer removal during Eukaryotic Okazaki fragment maturation Contributor Author Rossi, Marie Louise. Date Accessioned: 2009-02-23T17:05:09Z. Date Available: 2009-02-23T17:05:09Z. Date Issued: 2009-02-23T17:05:09Z. Identifier Uri: http://hdl.handle.net/1802/6537. Description: Dr. Robert A. Bambara, Faculty Advisor. Thesis (PhD) - School of Medicine and Dentistry, University of Rochester. UR only until January 2010. UR only until January 2010. G.Gibson, S.V Muse. A primer of genome science. 2nd ed. Sinauer. ISBN 0-87893-232-1 T A Brown. Gene Cloning and DNA analysis. 4nd Blackwell P. ISBN 0-632-05901-X
Retrieved from "http://en.wikipedia.org/"
|